Welcome to the Genomics and Molecular Epidemiology Research (GaMER) lab in the Department of Pathobiology and Veterinary Science of University of Connecticut.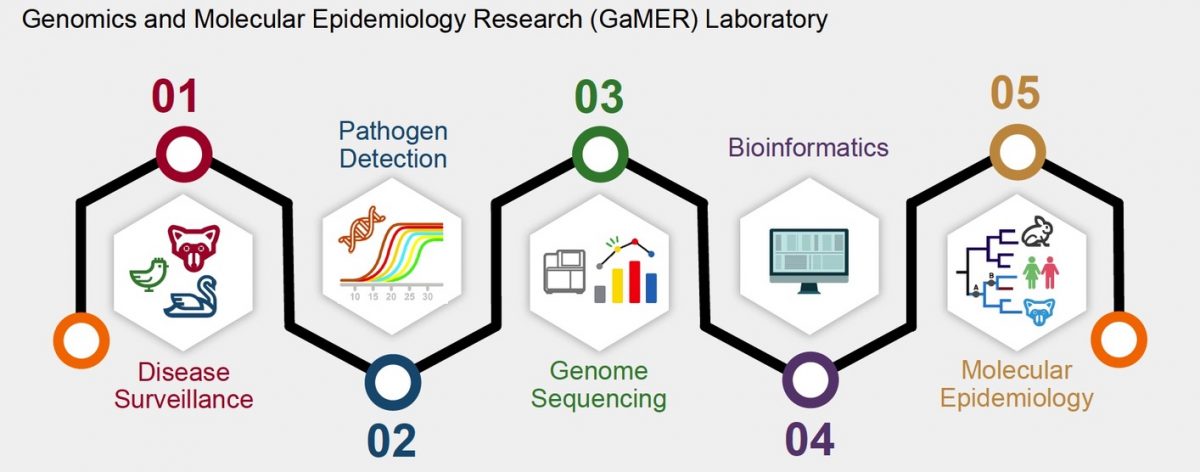
We use an interdisciplinary combination of high-throughput genome sequencing, computational biology, phylogenetic analysis, and traditional molecular biology and virology. We hope that, through our research, we can better understand the genetic diversity and evolutionary dynamics of rapidly mutating viruses in animals.
| Genome Sequencing | Tracking Virus | PVS4000 class |
News
- David`s new publication. Congratulations!Low Pathogenicity Avian Influenza (H5N2) Viruses, Dominican Republic Authors David H Chung, Dejelia R Gomez, Julia M Vargas, Belkis L Amador, Mia K Torchetti, Mary L Killian, David E Swayne, Dong-Hun Lee Publication date 2020/12 Journal Emerging infectious diseases Description Low pathogenicity avian influenza (H5N2) virus was detected in poultry in the Dominican Republic in […]
- David`s new paper. Congratulations!Evolution of H9N2 avian influenza viruses in Iran, 2017–2019 Authors Mohsen Bashashati, David H Chung, Mohammad Hossein Fallah Mehrabadi, Dong‐Hun Lee Publication date 2020/12/1 Journal Transboundary and Emerging Diseases Description Since its first detection in 1998, avian influenza virus (AIV) subtype H9N2 has been enzootic in Iran. To better understand the evolutionary history of H9N2 […]
- Sol Jeong`s new paper. Congratulations!Introduction of avian influenza A (H6N5) virus into Asia from North America by wild birds Authors Sol Jeong, Dong-Hun Lee, Yu-Jin Kim, Sun-Hak Lee, Andrew Y Cho, Jin-Yong Noh, Erdene-Ochir Tseren-Ochir, Jei-Hyun Jeong, Chang-Seon Song Publication date 2019/11 Journal Emerging infectious diseases Description An avian influenza A (H6N5) virus with all 8 segments of North […]
“A planning committee of the National Academies of Sciences, Engineering, and Medicine will organize a public workshop to explore research priorities for readiness and response to the Highly Pathogenic Avian Influenza outbreak in the United States.”
Genetic tracing at the Huanan Seafood market further supports an animal origin to #COVID19.
Learn more in @CellCellPress:
https://cell.com/cell/fulltext/S0092-8674(24)00901-2
@MichaelWorobey
@Sorbonne_Univ_ Florence Débarre
@scrippsresearch Kristian Andersen
'Virology—The next fifty years' - a perspective by Eddie Holmes, @GoodrumVirusLab and myself in Cell
https://www.cell.com/cell/fulltext/S0092-8674(24)00785-2
#H5N1 virus was detected in dead seals on Tyuleniy Island in eastern Russia, in the Sea of Okhotsk. Viruses isolated from the seals belong to clade 2.3.4.4b and are closely related to viruses detected in the Russian Far East and Japan in 2022–2023.
https://bit.ly/4elyZAF
Events at Uconn
Error loading calendar. Please make sure a valid RSS Feed is entered in the block settings.